
The same head imaged with different techniques
This is particularly challenging for MR images, as these show a weighted image of several tissue properties (in the most basis applications: T1, T2 and hydrogen density). How these properties are combined to a single image depends on the settings of the scanner.
MRIsim provides the student with a virtual MR scanner. He can set the scan parameters, and the program will produce the corresponding image for a real patient. In order to achieve this the program uses the underlying tissue parameters for these patients, obtained by interpolations from scans of that patient with different scan parameters.
By varying the scan parameters and observing the results, the student acquires an understand of the relation between the scan parameters and the underlying tissue parameters.
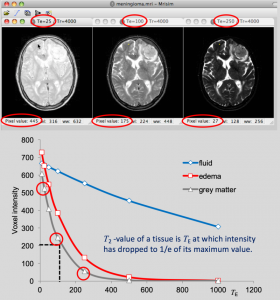
In this example, MRIsim is used to construct images for different values of TE. The student reads the signal intensity of pixels for different tissues as a function of TE, and makes a plot of the relation. In this way he discovers that the signal intensity drops with increasing TE, but with different rates for different tissues, discovering in this way the T2-property of the tissues
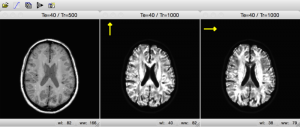
In another application, MRIsim uses gradient double-pulses in different directions. In this way the student discovers how this technique can be used to visualize diffusion within the tissue.
Currently MRIsim can produce spin-echo images, inverse recovery images, diffusion weighted images and fMRI images. New imaging techniques will be added in the future. User contributions, such as suggestions for additions or patient cases, are welcome.